Assessment of cardiac valve motion on time-resolved MRI images using deep learning
 System Overview
System Overview
The presented work aimed to develop a novel method for the task of valve motion assessment for prospective slice tracking in CMR temporal image (CINE) acquisition. The objective this thesis 1 was to evaluate deep learning-based landmarks tracking methods to extract the motion of the mitral valves throughout the cardiac cycle on time-resolved CMR four-chamber- view (4CHV) images. The fully automated CNN based algorithm would improve the current slice following acquisition workflow, which needs from manual user intervention. The automatic imaging slice tracking will enable a more precise morphology and flow estimation, potentially improving the diagnosis of diastolic dysfunction. During this project, an automatic deep learning algorithm using landmark detection was developed.
Motivation
Cardio-vascular diseases remain the leading cause of death worldwide [1]. In particular, diastolic dys- function is an important cause of cardiac insufficiency, heart failure, potentially leading to premature cardiovascular death if not diagnosed and treated for at an early disease stage. Left ventricular dias- tolic dysfunction is estimated to affect from 27% to 43% in middle-aged adults [2] and its prevalence increases with age.
Diastolic dysfunction is defined as a malfunctioning filling of the heart during diastole [3]. Its diag- nostic remains challenging as not only structural but functional abnormalities need to be evaluated [4]. Several non-invasive imaging techniques have been used for assessing diastolic dysfunction. Typically, intra-cardiac haemodynamics are measured using Doppler echocardiography. The peak velocities during early diastolic filling (E wave) and atrial contraction (A wave) are measured, and their ratio is calculated [5]. The Doppler flow measures are influenced by multiple factors including age, valve heart diseases, heart rate. Therefore, no single parameter is determinant enough to asses diastolic dysfunction [6].
Magnetic Resonance Imaging (MRI) is a noninvasive technique, well suited for disease diagnosis and monitoring due to its high spatial and temporal resolution and excellent soft-tissue contrast. In- deed, Cardiac Magnetic Resonance (CMR) provides information about heart structure and function, particularly in soft tissue characterization without the need of any contrast agent. Furthermore, CMR is useful for characterizing the anatomic valve morphology and cine images allows to visualize the valve throughout the cardiac cycle [7, 8]. Indeed, CMR allows assessment of blood flow using phase-contrast MRI (PC-MRI) [9].
As the valves move during a cardiac cycle, the acquisition of a fixed 2D slice will not allow the accurate time-resolved visualization of the valve and quantification of the blood flow: The valve moves into and out of the MRI slice so cannot be seen on each cardiac phase, therefore, the quantified flow through the valve is not correct. Despite the importance of correct valve flow assessment, time resolved CMR is yet usually performed at fixed slice positions throughout the cardiac cycle Kozerke et al. [10] introduced the prospective slice tracking concept. If the valve motion is known prior to the examina- tion, the slice position can be updated for each cardiac phase. As this approach however requires a dedicated pre-scan including an image-based algorithm to quantify the valve motion, very few related work can be found applying this technique in a clinical setting.
The recent development of deep learning has led to significant improvements in medical image analysis [11]. The prominent algorithms for feature tracking include deep learning systems based on convolutional neural network architectures. Specifically, for valve-tracking, a deep learning feature tracking algorithm for automatic slice-tracking can be developed, overcoming many limitations of the current valve-tracking techniques. The imaging slice tracking will enable a more precise morphology and flow estimation, potentially improving the diagnosis of diastolic dysfunction.
Dataset
The dataset is composed of Cardiac Magnetic Resonance Imaging 4CHV CINE series from 87 patients extracted from the Cardiac Atlas Project [Fon11]. The imaging protocol included CINE images acquired in long-axis planes, but for our work we only selected the 4CHV long axis view. The sequence of 4CHV CINE CMR are acquired on multiple 1.5T MRI scanners from different vendors (Philips, Siemens, GE). The mean pixel spacing of all selected datasets is 1.49 + 0.28 mm/pixel. Additionally the annotated dataset contains anatomical landmarks, i.e. the mitral valve position at each temporal aframe nnotated by an experienced analyst. As a preprocessing step, the CINE series were interpolated into a fixed and temporal resolution and horizontally flipped to have all the images oriented with the apex of the heart upwards. Finally,the input images intensity were normalized to 0-1 range values. After cleaning the data, the dataset contain 87 series that are split into 70% for training and the rest 30% equally between test and validation. To increase the variability of the dataset, online data augmentation was performed when training the network in forms of shift, center cropping, rotation, Guassian noise addition, contrast enhancement and Gaussian blurring.
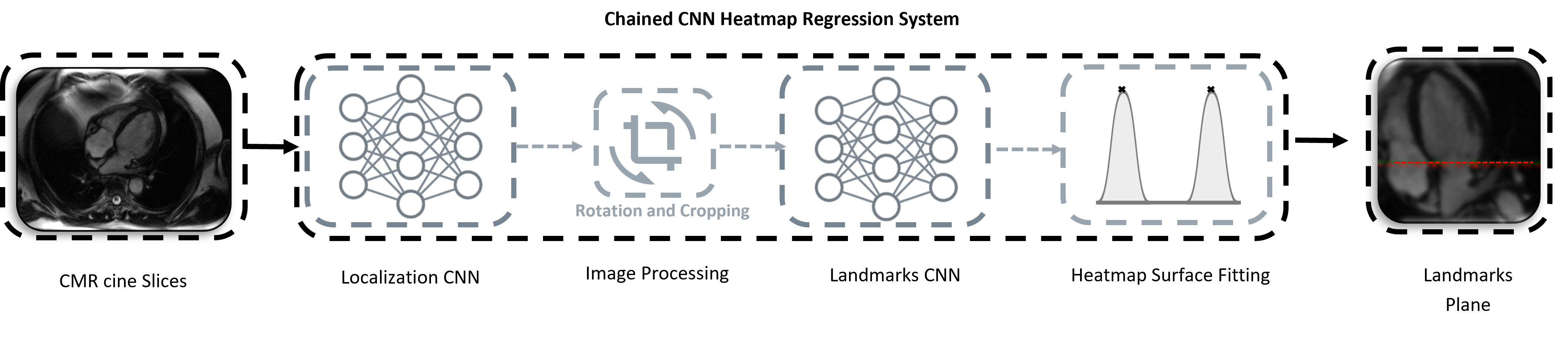
System Overview
The motion assesment system is composed of four main stages. The preprocessed CINE dataset are forwarded as input to a convolutional based neural network (CNN). The final proposed system comprises two chained CNNs based on heatmap regression approach: Localization Network + Landmark Detection Network. The task of the localization CNN model is to detect the landmarks only in the first temporal frame of the full 4CHV CINE series. The complete system is designed to regress the mitral valve-annulus landmarks for each time-frame points. Finally, the valvular plane motion can be derived from the two predicted landmark distance. The predicted temporal coordinates are translated into mm-space.
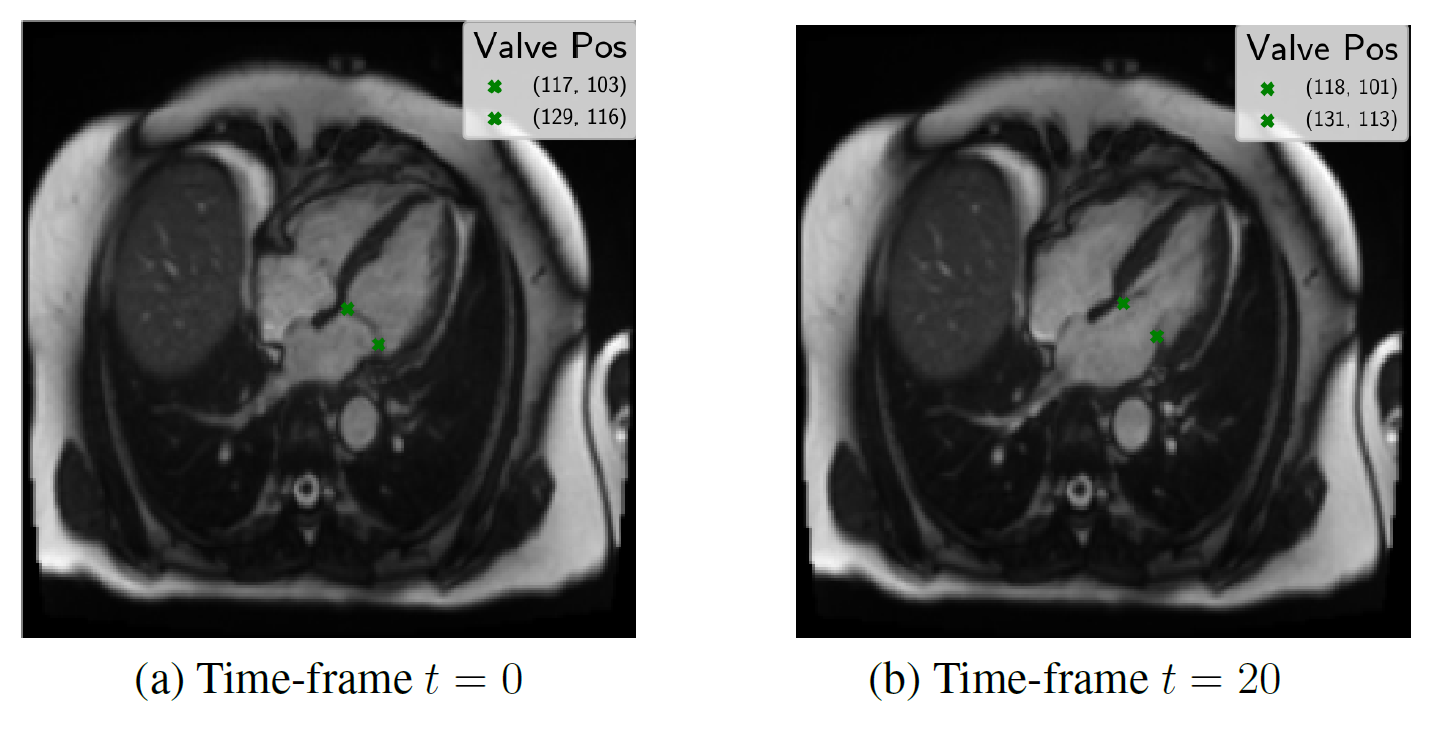
Results
Evaluated heatmap based regression approach for landmark detection yields to a superior performance than direct coordinate regression.Especially, the motion is better modeled by the two-stages architectures than with 3-D instead of single network. The proposed system showed an accurate match between expert-annotated and automatically detected landmarks for each time-frame. An example results of the model is shown in the below figure:
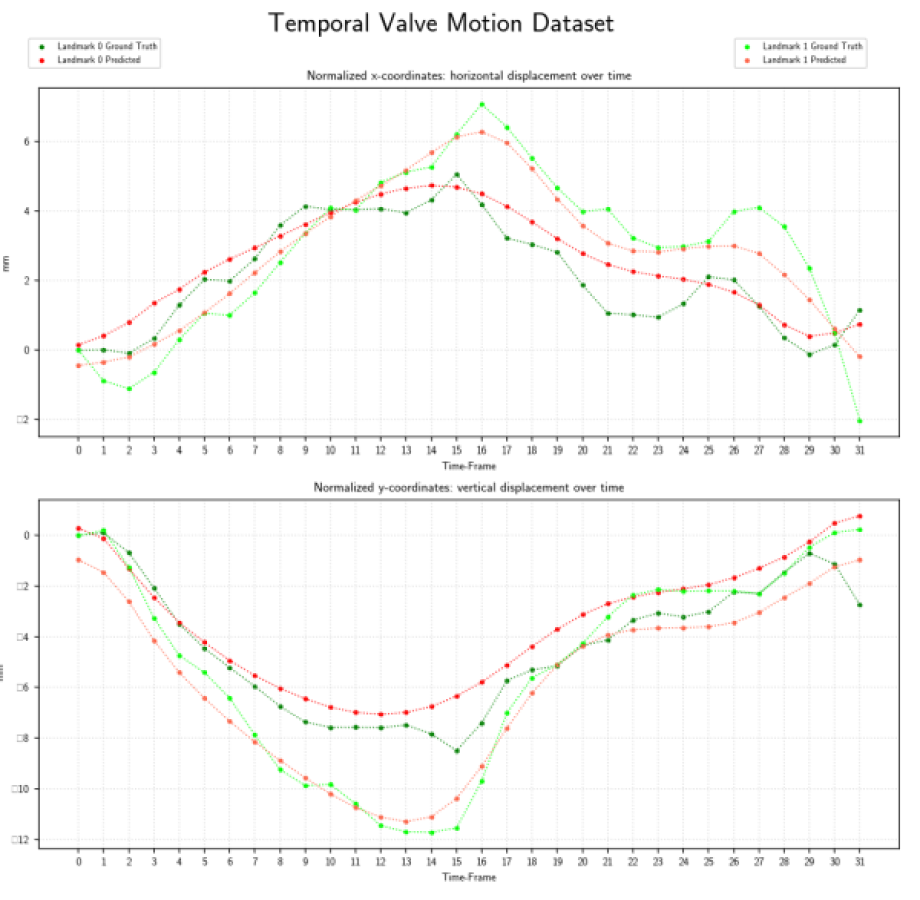
Our proposed method allows landmark localization with sub-pixel accuracy. Experiments show that our approach is able to correctly locate the landmark which can assess the prospective slice tracking acquisition method. The results of this work can be summarized as follows:
- CNNs can be used to develop a time-resolved landmark tracking application without the need of any user interaction.
- Heart landmark detection with the deep learning results can be improved by means of regressing heatmaps.
- The proposed two-stages method further allows a more accurate localization.
- A post-processing step with the non-linear least square fitting is able to refine the landmark location. The additional step outputs subpixel maxima and therefore predicts a smooth motion.
Conclusion and Outlook
The proposed system enables the valve tracking detection over time and therefore smooth valve motion assessment. Future work would focus on extension on the algorithm to 2CHV valve landmark detection. Evaluation with more test data and further refinement of the network. An integration of a single CNN could be convenient for faster inference time. Finally, when the validity of the method is proved , the scanner integration would be the next step.
References
[1] World Health Organization, World Health Statistics 2019: Monitoring Health for the SDGs. Geneva, Switzerland: World Health Organization, 2019.
[2] M. Nayor, L. L. Cooper, D. M. Enserro, V. Xanthakis, M. G. Larson, E. J. Benjamin, J. Aragam, G. F. Mitchell, and R. S. Vasan, “Left ventricular diastolic dysfunction in the community: Impact of diagnostic criteria on the burden, correlates, and prognosis,” Journal of the American Heart Association, vol. 7, no. 11, 2018.
[3] A. Kossaify and M. Nasr, “Diastolic dysfunction and the new recommendations for echocardiographic assessment of left ventricular diastolic function: Summary of guidelines and novelties in diagnosis and grading,” Journal of Diagnostic Medical Sonography, vol. 35, no. 4, pp. 317–325, 2019.
[4] C. Gutierrez and D. G. Blanchard, “Diastolic heart failure: Challenges of diagnosis and treatment,” American Family Physician, vol. 69, no. 11, pp. 2609–2616, 2004.
[5] C. Dugo, M. Rigolli, A. Rossi, and G. A. Whalley, “Assessment and impact of diastolic function by echocardiography in elderly patients.,” Journal of geriatric cardiology : JGC, vol. 13, no. 3, pp. 252–25260, 2016.
[6] S. F. Nagueh, O. A. Smiseth, C. P. Appleton, I. Byrd, Benjamin F., H. Dokainish, T. Edvardsen, F. A. Flachskampf, T. C. Gillebert, A. L. Klein, P. Lancellotti, P. Marino, J. K. Oh, B. Alexandru Popescu, and Waggoner, “Recom- mendations for the Evaluation of Left Ventricular Diastolic Function by Echocardiography: An Update from the American Society of Echocardiography and the European Association of Cardiovascular Imaging,” European Heart Journal - Cardiovascular Imaging, vol. 17, pp. 1321–1360, 07 2016.
[7] S. Shah, E. D. Chryssos, and H. Parker, “Magnetic resonance imaging: a wealth of cardiovascular information.,” The Ochsner journal, vol. 9, no. 4, pp. 266–77, 2009.
[8] K. Maganti, V. H. Rigolin, M. E. Sarano, and R. O. Bonow, “Valvular heart disease: diagnosis and management.,” Mayo Clinic proceedings, vol. 85, pp. 483–500, may 2010.
[9] P. Waheed, A. K. Naveed, and F. Farooq, “Cardiovascular magnetic resonance physics for clinicians: part I,” Journal of the College of Physicians and Surgeons Pakistan, vol. 19, no. 4, pp. 207–210, 2009.
[10] S. Kozerke, J. Schwitter, E. M. Pedersen, and P. Boesiger, “Aortic and mitral regurgitation: Quantification using moving slice velocity mapping,” Journal of Magnetic Resonance Imaging, vol. 14, no. 2, pp. 106–112, 2001.
[11] A. Maier, C. Syben, T. Lasser, and C. Riess, “A gentle introduction to deep learning in medical image processing,” Zeitschrift f ̈ur Medizinische Physik, vol. 29, no. 2, pp. 86 – 101, 2019.
[12] S. S. Yoon, E. Hoppe, M. Schmidt, C. Forman, P. Sharma, C. Tilmanns, A. Maier, and J. Wetzl, “Automatic Cardiac Resting Phase Detection for Static Cardiac Imaging Using Deep Neural Networks,” in Proceedings of the Joint Annual Meeting ISMRM-ESMRMB (27th Annual Meeting & Exhibition) (I. S. for Magnetic Resonance in Medicine, ed.), 2019.
-
All the algorithims will be developed in python using the Pytorch deep learning framework. The data for training, validation and testing will be provided by Siemens Healthineers. ↩︎